Introduction to Machine Learning in R
- Authors: Raul Samayoa (@ralle123)
- Lesson topic: Machine learning
- Lesson content URL: https://github.com/UofTCoders/studyGroup/tree/gh-pages/lessons/r/ml-intro
Slides are available at this link
Demo examples:
Linear regression
#install.packages("lattice")
library(lattice)
alligator <- data.frame(
lnLength = c(3.87, 3.61, 4.33, 3.43, 3.81, 3.83, 3.46, 3.76,
3.50, 3.58, 4.19, 3.78, 3.71, 3.73, 3.78),
lnWeight = c(4.87, 3.93, 6.46, 3.33, 4.38, 4.70, 3.50, 4.50,
3.58, 3.64, 5.90, 4.43, 4.38, 4.42, 4.25)
)
# Plot our information
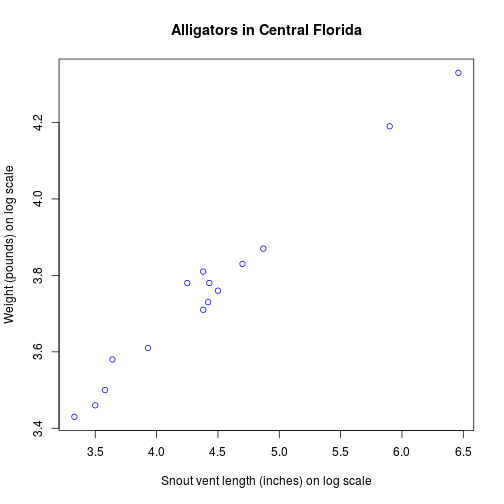
plot(
alligator$lnWeight,
alligator$lnLength,
col = "blue",
xlab = "Snout vent length (inches) on log scale",
ylab = "Weight (pounds) on log scale",
main = "Alligators in Central Florida"
)
lm function fits a linear model. A typical model has the form response ~ terms
myModel <- lm(lnLength ~ lnWeight, data = alligator)
Visualize the regression
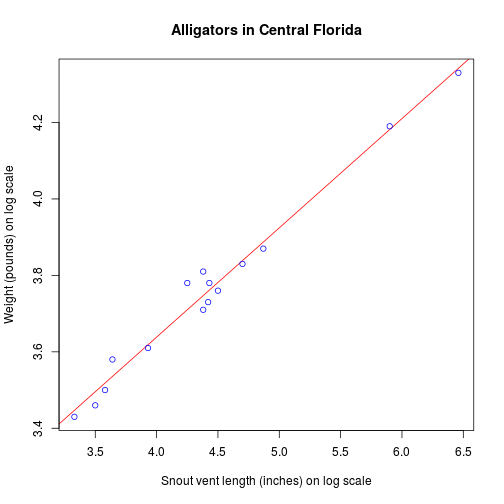
# Plot the chart with lm.
plot(
alligator$lnWeight,
alligator$lnLength,
col = "blue",
abline(myModel, col = 'red'),
xlab = "Snout vent length (inches) on log scale",
ylab = "Weight (pounds) on log scale",
main = "Alligators in Central Florida"
)
# Find weight of a alligator with weight of 5.5.
find_value <- data.frame(lnWeight = 5.5)
result <- predict(myModel, find_value)
result
## 1
## 4.067312
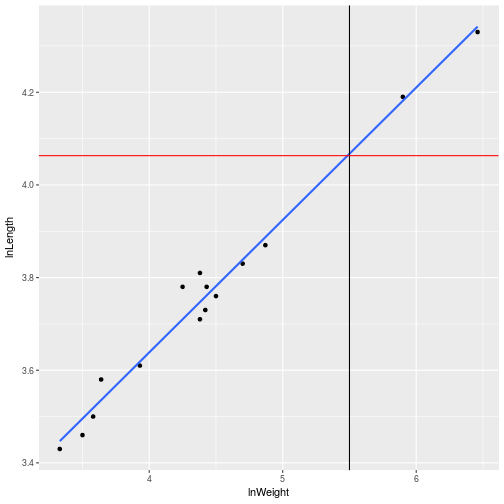
If we trace the line with the value we are looking for we can see that the value seems correct
library(ggplot2)
end <- ggplot(alligator, aes(x = lnWeight, y = lnLength)) +
geom_point()
end + geom_smooth(method = 'lm', se = FALSE) +
geom_vline(xintercept = 5.5) +
geom_hline(yintercept = 4.06312, color = 'red')
k-means
library(datasets)
head(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
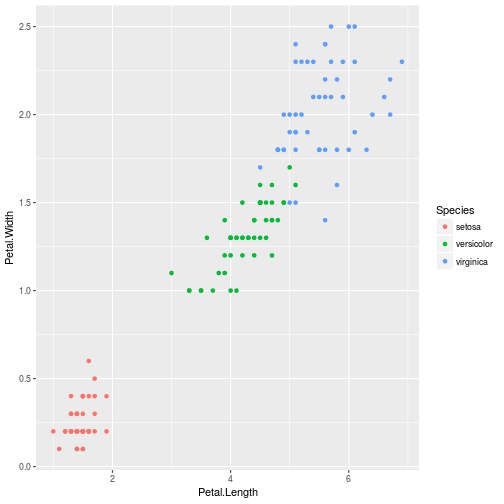
ggplot(iris, aes(Petal.Width, Petal.Length)) +
geom_point()
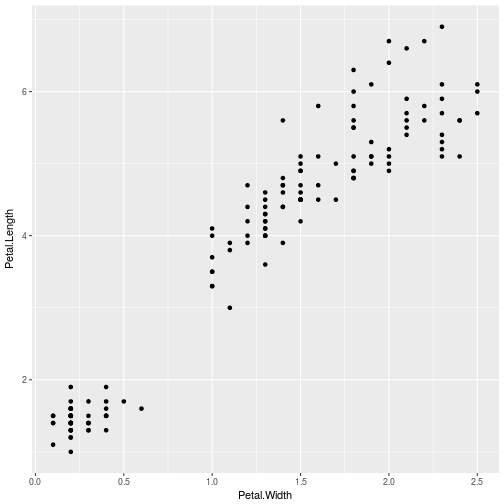
After resetting the seed I do a kmeans just selecting the 3-4 columns of iris, collect 3 iterations, selecting 20 random sets
set.seed(20)
irisCluster <- kmeans(iris[, 3:4], 3, nstart = 20)
#Comparing cluster with species
table(irisCluster$cluster, iris$Species)
##
## setosa versicolor virginica
## 1 50 0 0
## 2 0 48 4
## 3 0 2 46
We can graph our result
irisCluster$cluster <- as.factor(irisCluster$cluster)
ggplot(iris, aes(Petal.Length, Petal.Width, color = irisCluster$cluster)) +
geom_point()
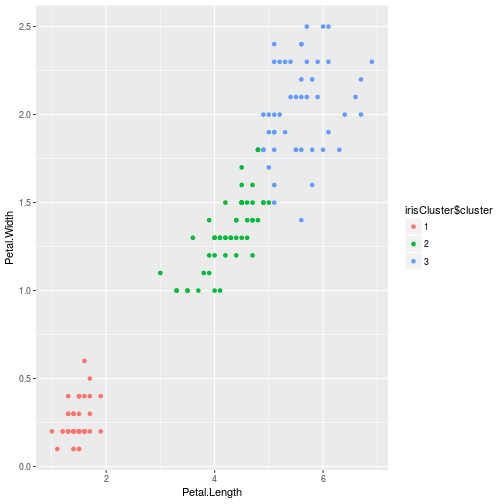
Why did we do 3 clusters and not 4? dataset only has 3 species
ggplot(iris, aes(Petal.Length, Petal.Width, color = Species)) +
geom_point()